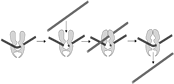Research
Research in the lab falls into the following themes:
-Biochemistry and Cell Biology of human DNA TOP2.
-Molecular Pharmacology and Genotoxicity of anti-DNA TOP2 drugs.
-Role of TOP2 in the aetiology of therapy related leukaemia.
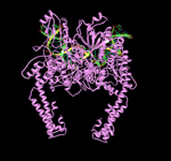
DNA TOP2, is essential for cell survival playing a role in replication, transcription, chromosome condensation, segregation and probably in overall chromatin domain organization. In vertebrates these activities are to some extent split between the two TOP2 paralogues, TOP2A and TOP2B, which are encoded by genes on chromosomes 17 and 3 , respectively [Austin & Fisher 1990]. TOP2B is the predominant form in quiescent and terminally differentiated cells, while TOP2A and TOP2B are both abundant in cycling cells [Padget, 2000]. The enzymatic activity of TOP2 allows one double-stranded DNA segment to pass through another, thus altering DNA topology. This is achieved via a TOP2-bridged double-strand break that is introduced into one DNA duplex. Each monomer of the dimeric enzyme remains covalently attached to the ends of the DSB via a 5’-phosphotyrosyl linkage. A second DNA segment can then be passed through the enzyme-bridged DNA gate, before the break is re-ligated (see diagram on the left). The crystal structure of the catalytic core of TOP2A bound to DNA has recently been solved [Wendorff et al 2012], adding to our understanding of the enzymes catalytic action. The TOP2-bridged gate is normally a transient intermediate, but there are a group of compounds that inhibit the religation step, resulting in the formation of an unusual type of DSB called a cleavage complex, in which the topoisomerase protein remains covalently coupled to the DNA. Compounds with this activity are called TOP2 poisons. The DNA lesions induced by TOP2 poisons are cytotoxic, hence the utility of these agents in cancer therapy. Unfortunately, TOP2 poisons are also associated with the formation of leukemogenic chromosome translocations [Cowell 2012].
A major part of our current work is supported by Bloodwise programme grant funding. We are investigating how anti-TOP2 drugs contribute to therapy related leukaemia [see Cowell and Austin for review] with the aim of identifying strategies that can reduce the rate of this devastating side effect, while maintaining the efficacy of current anti-cancer therapies. We have recently shown that TOP2B is disproportionatly responsible for the genotoxic properties of etoposide using a cell line model [Cowell et al 2012], and gathered evidence for a transcription-related mechanism for generating chromosome translocations [Cowell and Austin 2012].
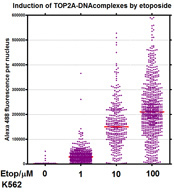
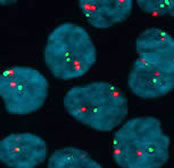
In order to quantify the specific type of DNA lesions introduced by TOP2 poisons such as etoposide (enzyme-linked TOP2-DNA complexes otherwise known as cleavage complexes) we use the Trapped in AgaRose DNA ImmunoStaining (TARDIS) assay which was developed in Newcastle. This method was recently reviewd by Cowell et al and we routinely use it to study the accumulation and processing of TOP2 DNA lesions after exposure to TOP2 poisons such as etoposide or dietary components [for example, Willmore et al 2004; Lopez-Lazaro et al 2010; Lee et al 2012].Other techniques routinely used in the lab include DNA and RNA FISH, immunofluorescence, ChIP, ChIP-seq and micronucleus asays
I co-discovered the TOP2B isoform [Austin and Fisher, 1990] and we can express recombinant human DNA TOP2A and TOP2B in quantity in a yeast expression system [Austin et al, 1995]. Mutated proteins have been used in structure-function studies [West et al, 2000; Leontiou et al, 2006].