caseparenttrios.dat: This file contains a header line (which is not read in by the program but is useful for reminding yourself of the column order), followed by a line of data for each of the 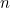 SNPs to be analysed (IN EXACTLY THE SAME ORDER as given in emimmarkers.dat).
SNPs to be analysed (IN EXACTLY THE SAME ORDER as given in emimmarkers.dat).
The first number on each line is the numeric SNP ID (as given in emimmarkers.dat). This is followed by 15 cell counts corresponding to the number of fully genotyped case/parent trios whose genotype combinations fall into the appropriate genotype categories as given in Ainsworth et al. (2011) Table 1. (Zero counts are allowed, although may make it more difficult to estimate certain parameter combinations).
For example, suppose that at the first SNP the genotype combinations of mother, father and child as given in Ainsworth et al. (2011) Table 1 are:
| group | mother | father | child | count |
|---|
| 1 | 22 | 22 | 22 | 4 |
| 2 | 22 | 12 | 22 | 10 |
| 3 | 22 | 12 | 12 | 17 |
| 4 | 12 | 22 | 22 | 9 |
| 5 | 12 | 22 | 12 | 13 |
| 6 | 22 | 11 | 12 | 6 |
| 7 | 11 | 22 | 12 | 4 |
| 8 | 12 | 12 | 22 | 14 |
| 9 | 12 | 12 | 12 | 44 |
| 10 | 12 | 12 | 11 | 26 |
| 11 | 12 | 11 | 12 | 25 |
| 12 | 12 | 11 | 11 | 17 |
| 13 | 11 | 12 | 12 | 24 |
| 14 | 11 | 12 | 11 | 12 |
| 15 | 11 | 11 | 11 | 19 |
Then the line in caseparenttrios.dat corresponding to this SNP would look like:
1 4 10 17 9 13 6 4 14 44 26 25 17 24 12 19
An example of caseparenttrios.dat for 8 SNPs, of which the first has counts as given above, is shown below:
snp cellcount 1-15
1 4 10 17 9 13 6 4 14 44 26 25 17 24 12 19
2 1 0 8 0 5 2 3 2 23 21 29 38 26 43 52
3 0 0 0 0 0 0 0 0 0 1 1 0 1 0 319
4 0 0 3 0 0 2 4 1 13 24 22 40 8 25 131
5 0 0 5 3 6 4 7 2 23 23 29 44 19 46 59
6 0 0 1 2 0 0 2 2 4 8 14 18 21 26 197
7 0 0 3 0 0 1 0 1 11 18 14 37 12 29 160
8 1 3 9 5 12 9 8 12 31 30 28 26 31 20 20