cases.dat: This file contains a header line (which is not read in by the program but is useful for reminding yourself of the column order), followed by a line of data for each of the 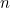 SNPs to be analysed (IN EXACTLY THE SAME ORDER as given in emimmarkers.dat).
SNPs to be analysed (IN EXACTLY THE SAME ORDER as given in emimmarkers.dat).
The first number on each line is the numeric SNP ID (as given in emimmarkers.dat). This is followed by 3 cell counts corresponding to the number of fully genotyped cases whose genotypes fall into the appropriate genotype categories. Note that these must not include cases already in the files caseparenttrios.dat, casemotherduos.dat, or casefatherduos.dat (i.e. all input data files must be independent).
For example, suppose that at the first SNP the genotypes of the cases are
| group | case | count |
|---|
| 1 | 22 | 93 |
| 2 | 12 | 268 |
| 3 | 11 | 174 |
Then the line in cases.dat corresponding to this SNP would look like:
1 93 268 174
An example of cases.dat for 8 SNPs, of which the first has counts as given above, is shown below:
snp cellcount 1-3
1 93 268 174
2 47 223 265
3 0 0 536
4 11 150 375
5 51 197 288
6 7 93 436
7 9 128 399
8 99 247 189