emimparams.dat: This is a file that tells EMIM what input files to read in and sets up the various parameter restrictions for the analyses to be performed. An example of this file is shown on the following page. Here we describe the lines of this file in detail:
Lines 1, 14 and 34 are not read by the program but are simply separators to make the 3 sections of this file easier to read. The text after 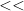 on each other line of the file is not read by the program, but is designed to describe what the number (1 or 0) at the beginning of the line means. You are strongly recommended to keep these text comments in order to avoid mistakes. The order of the lines must be EXACTLY as shown in this example.
on each other line of the file is not read by the program, but is designed to describe what the number (1 or 0) at the beginning of the line means. You are strongly recommended to keep these text comments in order to avoid mistakes. The order of the lines must be EXACTLY as shown in this example.
Lines 2-12 tell EMIM what input files to expect (note that the names given to these input files are not optional). A “1” indicates that this input file exists and is to be read in, while a “0” indicates that this input file is not to be read in. At least one of the files caseparenttrios.dat, casemotherduos.dat, casefatherduos.dat (lines 2, 4 and 5) must be read in; in the example above all three of these files are read in, as well as a conmotherduos.dat file.
Line 13 tells EMIM how many SNPs to be analysed: this number must either match or be less than the number of SNPs in the file emimmarkers.dat. (If this number n is less than the number of SNPs in the file emimmarkers.dat, then only the first n SNPs in the file emimmarkers.dat will be analysed).
The second and third sections of the file have a number of lines telling what parameters to estimate and what parameter restrictions to use. A “1” indicates that this parameter is to be estimated or this restriction is to be used. A “0” indicates that this parameter is not to be estimated or this restriction is not to be used.
-----------INPUT DATAFILES--------------------------------------
1 << caseparenttrios.dat file (0=no, 1=yes)
1 << caseparents.dat file (0=no, 1=yes)
1 << casemotherduos.dat file (0=no, 1=yes)
0 << casefatherduos.dat file (0=no, 1=yes)
1 << casemothers.dat file (0=no, 1=yes)
0 << casefathers.dat file (0=no, 1=yes)
1 << cases.dat file (0=no, 1=yes)
1 << conparents.dat file (0=no, 1=yes)
0 << conmotherduos.dat file (0=no, 1=yes)
0 << confatherduos.dat file (0=no, 1=yes)
1 << cons.dat file (0=no, 1=yes)
634 << no of SNPs in each file
------------------PARAMETER RESTRICTIONS-----------------------
0 << fix allele freq A (0=no, 1=yes)
1 << assume HWE and random mating (0=no=estimate mews, 1=yes)
0 << assume parental allelic symmetry (0=no, 1=yes)
0 << use CPG likelihood (9 mews)
1 << estimate R1 (0=no, 1=yes)
1 << estimate R2 (0=no, 1=yes)
0 << R2=R1 (0=no, 1=yes)
0 << R2=R1squared (0=no, 1=yes)
0 << estimate S1 (0=no, 1=yes)
0 << estimate S2 (0=no, 1=yes)
0 << S2=S1 (0=no, 1=yes)
0 << S2=S1squared (0=no, 1=yes)
1 << estimate Im (0=no, 1=yes)
0 << estimate Ip (0=no, 1=yes)
0 << estimate gamma11 (0=no, 1=yes)
0 << estimate gamma12 (0=no, 1=yes)
0 << estimate gamma21 (0=no, 1=yes)
0 << estimate gamma22 (0=no, 1=yes)
0 << gamma22=gamma12= gamma21=gamma11 (0=no, 1=yes)
---------------OTHER PARAMETERIZATIONS-------------------------
0 << estimate Weinberg (1999b) Im (0=no, 1=yes)
0 << estimate Weinberg (1999b) Ip (=Li 2009 Jm) (0=no, 1=yes)
0 << estimate Sinsheimer (2003) gamma01 (0=no, 1=yes)
0 << estimate Sinsheimer (2003) gamma21 (0=no, 1=yes)
0 << estimate Palmer (2006) match parameter (0=no, 1=yes)
0 << estimate Li (2009) conflict parameter Jc (0=no, 1=yes)
Line 15 << fix allele freq A (0=no, 1=yes) indicates that the allele frequencies are to be fixed at their given starting values (NOT RECOMMENDED). A “1” in this line will supercede any instructions given in the next two lines (lines 16 and 17).
Line 16 << assume HWE and random mating (0=no=estimate mews, 1=yes) indicates that the analysis should be performed assuming Hardy Weinberg Equilibrium (HWE) and random mating. In that case, one allele frequency parameter 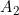 (the frequency of the 2 allele) will be estimated (or fixed) as opposed to estimating six mating-type stratification parameters
(the frequency of the 2 allele) will be estimated (or fixed) as opposed to estimating six mating-type stratification parameters 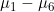 . A “1” in this line will supercede any instructions given in the next line (line 17).
. A “1” in this line will supercede any instructions given in the next line (line 17).
Line 17 << assume parental allelic symmetry (0=no, 1=yes) indicates that parental allelic symmetry should be assumed (i.e. 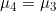 ) when estimating
) when estimating 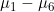 .
.
Line 18 << use CPG likelihood (9 mews) indicates that the Conditional on Parental Genotypes (CPG) rather than the Conditional on Exchangeable Parental Genotypes (CEPG) [Cordell (2004), Weinberg and Shi (2009)] likelihood should be used. This provides a more robust analysis that does not assume exchangeability of parental mating types, at the expense of estimating a larger number (nine) of mating-type stratification parameters. This analysis is recommended if your derives from pedigrees that contain (or were ascertained on the basis of the presence of) multiple affected individuals [Cordell (2004)].
Line 19 << estimate R1 (0=no, 1=yes) indicates that the child genotype effect 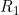 (the factor by which the disease risk is multiplied if the child has a single copy of allele 2) should be estimated.
(the factor by which the disease risk is multiplied if the child has a single copy of allele 2) should be estimated.
Line 20 << estimate R2 (0=no, 1=yes) indicates that the child genotype effect 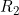 (the factor by which the disease risk is multiplied if the child has two copies of allele 2) should be estimated.
(the factor by which the disease risk is multiplied if the child has two copies of allele 2) should be estimated.
Line 21 << R2=R1 (0=no, 1=yes) indicates that a single child genotype effect 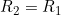 should be estimated. A “1” in this line will supercede any instructions given in the two previous lines. However, if line 21 is set equal to “1”, we recommend you set lines 19 and 20 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
should be estimated. A “1” in this line will supercede any instructions given in the two previous lines. However, if line 21 is set equal to “1”, we recommend you set lines 19 and 20 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
Line 22 << R2=R1squared (0=no, 1=yes) indicates that a single child genotype effect 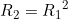 should be estimated. This is a multiplicative allelic model for the child genotype effects. A “1” in this line will supercede any instructions given in the three previous lines. However, if line 22 is set equal to “1”, we recommend you set lines 19, 20 and 21 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
should be estimated. This is a multiplicative allelic model for the child genotype effects. A “1” in this line will supercede any instructions given in the three previous lines. However, if line 22 is set equal to “1”, we recommend you set lines 19, 20 and 21 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
Line 23 << estimate S1 (0=no, 1=yes) indicates that the maternal genotype effect 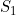 (the factor by which the disease risk is multiplied if the mother has a single copy of allele 2) should be estimated.
(the factor by which the disease risk is multiplied if the mother has a single copy of allele 2) should be estimated.
Line 24 << estimate S2 (0=no, 1=yes) indicates that the maternal genotype effect 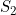 (the factor by which the disease risk is multiplied if the mother has two copies of allele 2) should be estimated.
(the factor by which the disease risk is multiplied if the mother has two copies of allele 2) should be estimated.
Line 25 << S2=S1 (0=no, 1=yes) indicates that a single maternal genotype effect 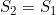 should be estimated. A “1” in this line will supercede any instructions given in the two previous lines. However, if line 25 is set equal to “1”, we recommend you set lines 23 and 24 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
should be estimated. A “1” in this line will supercede any instructions given in the two previous lines. However, if line 25 is set equal to “1”, we recommend you set lines 23 and 24 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
Line 26 << S2=S1squared (0=no, 1=yes) indicates that a single maternal genotype effect 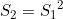 should be estimated. This is a multiplicative allelic model for the maternal genotype effects. A “1” in this line will supercede any instructions given in the three previous lines. However, if line 26 is set equal to “1”, we recommend you set lines 23, 24 and 25 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
should be estimated. This is a multiplicative allelic model for the maternal genotype effects. A “1” in this line will supercede any instructions given in the three previous lines. However, if line 26 is set equal to “1”, we recommend you set lines 23, 24 and 25 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
Line 27 << estimate Im (0=no, 1=yes) indicates that a maternal imprinting effect 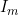 (a multiplicative factor by which the probability of disease is multiplied if the child receives a (maternal) copy of the 2 allele from their mother) should be estimated. A “1” in this line will supercede any instructions given in the next line (line 28) i.e. only one of
(a multiplicative factor by which the probability of disease is multiplied if the child receives a (maternal) copy of the 2 allele from their mother) should be estimated. A “1” in this line will supercede any instructions given in the next line (line 28) i.e. only one of 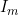 and
and 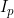 can be estimated. The exception to this is if no child genotype (R) or interaction (gamma) parameters are estimated, in which case it is possible to estimate both
can be estimated. The exception to this is if no child genotype (R) or interaction (gamma) parameters are estimated, in which case it is possible to estimate both 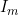 and
and 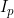 .
.
Line 28 << estimate Ip (0=no, 1=yes) indicates that a paternal imprinting effect 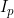 (a multiplicative factor by which the probability of disease is multiplied if the child receives a (paternal) copy of the 2 allele from their father) should be estimated.
(a multiplicative factor by which the probability of disease is multiplied if the child receives a (paternal) copy of the 2 allele from their father) should be estimated.
Line 29 << estimate gamma11 (0=no, 1=yes) indicates that the mother/child genotype interaction parameter 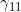 should be estimated.
should be estimated.
Line 30 << estimate gamma12 (0=no, 1=yes) indicates that the mother/child genotype interaction parameter  should be estimated.
should be estimated.
Line 31 << estimate gamma21 (0=no, 1=yes) indicates that the mother/child genotype interaction parameter 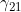 should be estimated.
should be estimated.
Line 32 << estimate gamma22 (0=no, 1=yes) indicates that the mother/child genotype interaction parameter 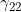 should be estimated.
should be estimated.
Line 33 << gamma22=gamma12=gamma21=gamma11 (0=no, 1=yes) indicates that a single mother/child genotype interaction parameter 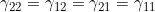 should be estimated. A “1” in this line will supercede any instructions given in the four previous lines. However, if line 33 is set equal to “1”, we recommend you set lines 29, 30, 31 and 32 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
should be estimated. A “1” in this line will supercede any instructions given in the four previous lines. However, if line 33 is set equal to “1”, we recommend you set lines 29, 30, 31 and 32 to “0” in order to avoid problems when EMIM tries to determine whether the parameters you have selected are estimable, given the data.
Depending on what optional input data files are available, estimation of certain parameter combinations may be limited. (This is particularly true if you only read in a single file, casemotherduos.dat or casefatherduos.dat ). EMIM will attempt to adjust the number of parameters to estimate in some “sensible” way if it detects you are trying to estimate too many parameters with not enough restrictions. However, it may be better to make this adjustment yourself (e.g. by making assumptions of HWE and/or estimating only a smaller number of parameters). You can generally tell if EMIM has been successful at its choice of parameters by looking at the output confidence intervals: if these do not look sensible (e.g. if the upper and lower confidence limits for a parameter are equal) then there is a good chance that the choice of parameters has not been made appropriately.
Lines 35 and 36 << estimate Weinberg (1999b) Im (0=no, 1=yes) << estimate Weinberg (1999b) Ip (=Li 2009 Jm) (0=no, 1=yes)
Parameterization of interactions and imprinting effects is quite complex (see Ainsworth et al. (2011)) and several different parameterizations have been proposed in the literature. Our paramaterization for the parent-of-origin effects 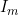 and
and 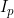 corresponds to the original parameterization used by Weinberg et al. (1998) rather than to a later alternative parameterization used by Weinberg (1999), Parimi et al. (2008), and Li et al. (2009). If preferred, the user can choose to use the later parameterization by setting the values in lines 27 and 28 to 0 and the values in line 35 or 36 to 1. In this case, if interactions are also required, we recommend using either the Sinsheimer et al. (2003) or Palmer et al. (2006) parameterization (see below), as our interaction parameterization does not allow estimation of the later Weinberg (1999) imprinting parameters.
corresponds to the original parameterization used by Weinberg et al. (1998) rather than to a later alternative parameterization used by Weinberg (1999), Parimi et al. (2008), and Li et al. (2009). If preferred, the user can choose to use the later parameterization by setting the values in lines 27 and 28 to 0 and the values in line 35 or 36 to 1. In this case, if interactions are also required, we recommend using either the Sinsheimer et al. (2003) or Palmer et al. (2006) parameterization (see below), as our interaction parameterization does not allow estimation of the later Weinberg (1999) imprinting parameters.
Lines 37 and 38 << estimate Sinsheimer (2003) gamma01 (0=no, 1=yes) << estimate Sinsheimer (2003) gamma21 (0=no, 1=yes) Sinsheimer et al. (2003) proposed an alternative parameterization for interactions in terms of maternal-fetal genotype incompatibility (MFG) parameters. Sinsheimer et al. (2003) denoted these parameters as 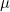 (or
(or 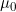 ) and
) and 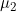 . We denote these MFG interactions as
. We denote these MFG interactions as 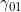 and
and 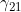 , since they correspond to effects that operate (in addition to maternal and child genotype effects) when the child has one copy, and the mother either zero or two copies, of a particular allele of interest. To include one or both MFG interactions, you should set the values in lines 29-33 to 0 and the value(s) in line 37 and/or 38 to 1.
, since they correspond to effects that operate (in addition to maternal and child genotype effects) when the child has one copy, and the mother either zero or two copies, of a particular allele of interest. To include one or both MFG interactions, you should set the values in lines 29-33 to 0 and the value(s) in line 37 and/or 38 to 1.
Line 39 << estimate Palmer (2006) match parameter (0=no, 1=yes) Sinsheimer et al. (2003) and Palmer et al. (2006) considered an alternative interaction parameterization in which “matching” rather “mismatching” between maternal and foetal genotypes increases disease risk in the offspring. To model interaction via the single Palmer et al. (2006) match parameter 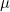 , you should set the values in lines 29-33 to 0 and the value in line 39 to 1.
, you should set the values in lines 29-33 to 0 and the value in line 39 to 1.
Line 40 << estimate Li (2009) conflict parameter Jc (0=no, 1=yes) Li et al. (2009) (based on work by Parimi et al. (2008)) considered an alternative interaction parameterization that modelled “conflict” between the mothers and childs genotypes. To model interaction via the single Li et al. (2009) conflict parameter (which we denote 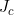 , corresponding to exp(
, corresponding to exp(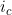 ) in the notation of Li et al. (2009)), you should set the values in lines 29-33 to 0 and the value in line 40 to 1. Note that the recommended model of Li et al. (2009) and Parimi et al. (2008)) is to include both
) in the notation of Li et al. (2009)), you should set the values in lines 29-33 to 0 and the value in line 40 to 1. Note that the recommended model of Li et al. (2009) and Parimi et al. (2008)) is to include both 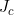 and
and 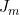 (=exp(
(=exp(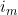 ) in the notation of Li et al. (2009)) where
) in the notation of Li et al. (2009)) where 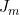 is the imprinting parameter selectable on line 36. So to fit the full Li et al. (2009) and Parimi et al. (2008) model you should set the values in lines 36 and 40 to 1.
is the imprinting parameter selectable on line 36. So to fit the full Li et al. (2009) and Parimi et al. (2008) model you should set the values in lines 36 and 40 to 1.